First, let’s begin by clarifying some terminology. As it pertains to taxonomy or identification, it is important to specify to what level of identification is required. The time-honored system of Linnaean classification (See Figure 1) is still our basis for microbial taxonomy or identification.
As we will see, in most cases for bacterial identification, our good friend the 16S gene sequence analysis, while not for all cases, is normally well suited to obtain a species name.14 What about use of the term “strain”? In some cases, “strain” has a dual meaning. It can refer just about to any organism added to a product. However, the term “strain” is frequently misused as synonomous with species. For example, “This is a three strain product” means it usually in informal language means that it contains three different species of probiotic microorganisms. In the taxonomic sense, a strain represents a subtype or subspecies ranking. Technically, if correctly classified, a strain is but a different member of the same species group. This is highly relevant biologically but still sounds picky, doesn’t it? Perhaps, but it is always necessary to be able to assure any label claims and at this point in time, we are still using the Linnaean system of nomenclature for nomenclature. Strain or subspecies names might be based solely on extrinsic factors like the name of the person who isolated the strain, or the number of the colony picked from a culture plate, or the source, a place, the date, etc. It also might be based on intrinsic biological traits (e.g., a specific trait or single base pair difference in an important gene of the species or even distinguishing a pathogen from its non-pathogemnic cousin).15
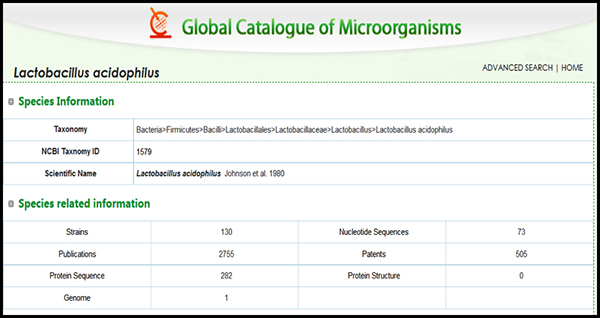
In an example (Figure 2), we see that there are 130 strains deposited in culture collections that are all speciated as Lactobacillus acidophoilus. In order to get to the “strain“ level of classification one must have a unique identifier that is a signature for any particular strain. That identifier may be as simple as knowing the source or much more complex as in knowing some signature phenotypic trait (colony or cellular morpholopgy, or some unique biochemical reaction for instance) or even more complex by a known DNA sequence difference as small as a single nucleotide difference (called a SNP or single nucleotide polymorphism).
Is it possible that several probiotic product producers are using the same strain of a particular probiotic? Yes, and in fact it is likely that strains available from just a few probiotic manufacturers are used throughout the probiotic industry under different labels and commercialized names. When asking for analysis, before using the term “strain” ask yourself if you understand exactly what it is you need. If it is a species name, then ask for species. If indeed you need to know if it is the exact same strain that was purchased from a manufacturer, then inquire to the manufacturer whether they already have an assay (such as a PCR-based test or a flow cytometry probe) specific for their strain or if they have had the strain sequenced and possess a whole genome sequence. If the former, an appropriate PCR assay can be used to detect to the strain level. If the later, the whole genome sequence data could be used to develop a PCR assay (which normally means contracting with a genomics lab for PCR primer and assay design) that can distinguish a strain-identifying marker.3,5 Alternatively, the sequence data could possibly be the basis of a flow cytometry probe for single strain compositions of probiotics.9,11
What About Mixed Probiotic Products?
Let’s move on to even more difficult determinations—i.e., analyzing probiotic mixtures of multiple species and strains. Again, it is not impossible that selective/differential agar media can separate and quantify to some level of resolution; but what that level is, other than the common descriptive or functional group name (e.g., some type of lactic acid bacterium, or some type of bifidobacteria) is unknown. If we assume the concentrated product is labeled with a genus and species name, can we get to the level of species or strain using culture? Yes, but, not directly. This is not only a long and tedious task, but a highly costly one. Assume we picked some representative number of colonies, say 5 to 10, from a selective/differential agar plate (that we chose because of the stated composition of the probiotic product), and streak for isolation and then proceed to have each colony individually identified by either a phenotypic or genomic method. For this relatively simple exercise, we are already amassing a bill of around $500–$1,000, and that is for just a single selective/differential agar plate! If a probiotic mixture requires two to four different media, then we jump to a conservative estimate of $1,000–$2,000 to get a name. Even then, all of this relies on a microorganism’s ability to form a colony under the growth conditions used as well as the lab technician subjectively picking a representative sampling of the different visible colony types. Although there are some amazingly specific selective/differential media available, these rarely can reliably get to the species level within the complex taxonomy of the Lactobacillaceae or Bifidobacteriaceae families. What culture can clearly do better than almost any other method is to enumerate colony forming units when the identity of the bacterium in a single-organism product is known and it is cultivable. Indeed, the standard recommended by the IPA (International Probiotics Association) is the CFU. To complicate matters further, probiotic marketing managers engage in a “more is better” product differentiation arms race. Where once you could market a probiotic with a single strain, it is now fashionable to add multiple strains to a product mix and to upsell products based on probiotic dose. Products with multiple probiotic strains and larger declared probiotic populations command a greater price than products with fewer strains and smaller populations. We dare you to try and verify such claims. It is easy to throw up one’s hands and ask, “this is all a big mess, what can we do?”
Fear not, take a deep breath, sit back and relax, because this is where you must agree that it is a great time to be a microbiologist. Now we are equipped with not only good cultural tests, but with the tools of genomics, specifically using the microbiome technique, we can determine the makeup of a mixture of probiotics without the need to culture, hope things grow, pick and then identify. Recall from the first article in this series that a microbiome is the identification of a community of bacteria in a single sample (see: Microbiomes Move Standard Plate Count One Step Forward and Metagenomes and Their Utility). We can perform a DNA extraction on the mixture and then using a 16S Targeted Bacterial Microbiome, resolve the bacterial mixture into a genus-level profile. Or, if the 16S sequence reads are of sufficient length, we can identify to the species level as well as determine the relative proportions of each taxon. If the probiotic mixture contains both bacteria and yeasts or other fungi, we can perform two different targeted microbiomes (bacterial and fungal), or rely on a metagenome-based microbiome to determine sequences and identify all microorganisms with signature DNA in that sample. If relative proportions of the constituents are not needed, we have an option to use a microbial microarray, which will give species names and in some cases even a strain or subtype. There are pros and cons to each method, but cost and very often time is saved with these approaches.
Finally, we would like to address issues of genomic methods to distinguish viability. As stated in a previous column, PCR-based methods detect DNA (or RNA) sequences and the quantity or copy numbers of sequences in a sample. Recall that PCR is similar in function to a copy machine in that it spits out billions of copies of a single gene sequence, with a final copy number that is proportional to the starting number. However, DNA in the sample could have been derived from dead (non-viable) cells. Because PCR counts genes, such dead cell genes, DNA can be detected resulting in a false identification. Is this a deal breaker for using genomic methods for microbial analysis of probiotics? No, not at all. There are assays that can use viability dyes (e.g., propidium monoazide) to distinguish live from dead bacteria.11 These dye assays can function well in both a PCR format or flow cytometry. Flow cytometry is currently receiving widespread attention because of assay speed and apparent ability to distinguish live, dead and sub-lethally injured cells.9,12,13
Knowledge is Power
Knowledge of your probiotic strain(s) is critical for quality assurance and regulatory compliance. Perhaps the ultimate in genomic characterization is to determine the whole genome sequence (WGS) of your organism(s).15,16,17 This sequence provides many potential uses:
- Sequence sites for designing strain-specific PCR or flow cytometry primers and probes
- Product mix identifications for regulatory compliance
- Assurance of strain ID for customers
- Presence of single nucleotide polymorphisms for intellectual property protection
- Quality control assays for maintaining stock and production strain purity
- Assurance that strains are non-pathogenic subtypes of species that might have pathogenic strains within the same species
- Assessing the presence of functional genes that confer beneficial (digestion aids, gut survival) or detrimental (toxins or virulence factors, antibiotic resistance) characteristics to a strain.
It must be stated that this science is still very much in development, and which methods might become more standard across the industry is still a wide open and rapidly evolving question. For example, there are issues with the accuracy and strain-resolution capability of these methods. One group reported that probiotic compositions often do not meet the stated label claims.18 These authors pointed out that of 52 different probiotic products tested, one-third were below label claim for cell counts (CFU) before expiration. They further demonstrated by using a multiplexed-PCR assay (a PCR assay that detects more than a single target in a single assay) that 58% of the sample set had incorrect labels, including:
- The name of the organism due to wrong taxonomy
- Undeclared species were found that were not on the label
- Species on the label were missing from the product.
Indeed, the science of subtyping, which is not an easy task in part due to the very complex family structures of many microbial groups, as well as their less-than-hardy constitution, needs much improvement. For example, one report demonstrated that only a single example out of 16 Bifidobacterium spp. probiotic preparations correctly matched the label claims for viability and species or subspecies type.19
The combination of culturing with some form of genomic analysis is a powerful tool that probiotic manufacturers should exploit, and we hope this column has helped by sharing some basic information. We have covered a smattering of genomic methods—these are by no means the only ones, but we have selected tools that are currently gaining favor for their precision and accuracy over other methods. We also see that genomic tools are not only useful for probiotic producers and users, but they are becoming the standard for probiotic analysis by regulators. Regulatory science and agencies currently rely upon the level of detail provided by genomic methods to help support their activities.
What is in the near future of this field? Look for suggestions that other genes besides the 16S gene will be used to discern and resolve complicated taxonomic groups. Look for multiplexed PCR assays coupled with viability indicators to become in more widespread use. Finally, look for rapid whole genome sequencing tools for developing probes specific for not only species but also your own specific strains. Such tools will become significantly less costly and more rapid than current reality. The future in this arena is quite bright.
In this installment of Food Genomics we explored the basis and some uses of genomic tools for probiotic analysis. We hope the reader is now more prepared to ask the right questions and understand which tools are available for which job. As always, please email us with your comments and questions.
References
- Global Probiotics Market to Reach $24 Billion by 2017. (December 16, 2013). Retrieved from https://www.naturalproductsinsider.com/news/2013/12/global-probiotics-market-to-reach-24-billion-by-2.aspx
- Probiotics in Animal Feed Market Worth 5.07 Billion USD by 2022 | Markets Insider. Accessed November 15, 2017. Retrieved from http://markets.businessinsider.com/news/stocks/Probiotics-in-Animal-Feed-Market-Worth-5-07-Billion-USD-by-2022-1002272597
- Treven, P. (2015). Strategies to develop strain-specific PCR based assays for probiotics. Beneficial Microbes, 6(6), 887–898. https://doi.org/10.3920/BM2015.0009
- Guidelines for Probiotics in food. (n.d.). Accessed November 15, 2017. Retrieved from http://internationalprobiotics.org/resources/guidelines/
- Huys, G., Botteldoorn, N., Delvigne, F., Vuyst, L. D., Heyndrickx, M., Pot, B., Daube, G. (2013). Microbial characterization of probiotics–Advisory report of the Working Group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Molecular Nutrition & Food Research, 57(8), 1479–1504. https://doi.org/10.1002/mnfr.201300065
- Parte, A.C. (n.d.). LPSN – List of Prokaryotic names with Standing in Nomenclature. Retrieved from http://www.bacterio.net/index.html
- Davis, C. (2014). Enumeration of probiotic strains: Review of culture-dependent and alternative techniques to quantify viable bacteria. Journal of Microbiological Methods, 103(Supplement C), 9–17. https://doi.org/10.1016/j.mimet.2014.04.012
- Daniels, S. (n.d.). IPA petitions FDA on requiring CFUs for probiotic labeling. Accessed November 15, 2017. Retrieved from https://www.foodnavigator-usa.com/Article/2016/11/22/IPA-petitions-FDA-on-requiring-CFUs-for-probiotic-labeling
- Bunthof, C. J., & Abee, T. (2002). Development of a Flow Cytometric Method To Analyze Subpopulations of Bacteria in Probiotic Products and Dairy Starters. Applied and Environmental Microbiology, 68(6), 2934–2942. https://doi.org/10.1128/AEM.68.6.2934-2942.2002
- Sattler, V. A., Mohnl, M., & Klose, V. (2014). Development of a strain-specific real-time PCR assay for enumeration of a probiotic Lactobacillus reuteri in chicken feed and intestine. PloS One, 9(2), e90208. https://doi.org/10.1371/journal.pone.0090208
- Fittipaldi, M., Nocker, A., & Codony, F. (2012). Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. Journal of Microbiological Methods, 91(2), 276–289. https://doi.org/10.1016/j.mimet.2012.08.007
- Kramer, M., Obermajer, N., Bogovic Matijasić, B., Rogelj, I., & Kmetec, V. (2009). Quantification of live and dead probiotic bacteria in lyophilised product by real-time PCR and by flow cytometry. Applied Microbiology and Biotechnology, 84(6), 1137–1147. https://doi.org/10.1007/s00253-009-2068-7
- ISAPP. (2016, January 4). Flow Cytometry for Probiotic Enumeration | ISAPP. Retrieved from https://isappscience.org/flow-cytometry-for-probiotic-enumeration/
- Hug, L. A., Baker, B. J., Anantharaman, K., Brown, C. T., Probst, A. J., Castelle, C. J., Banfield, J. F. (2016). A new view of the tree of life. Nature Microbiology, 1(5), 16048. https://doi.org/10.1038/nmicrobiol.2016.48
- Natarajan, P., & Madasamy, P. (2014). First Complete Genome Sequence of a Probiotic Enterococcus faecium Strain T-110 and Its Comparative Genome Analysis with Pathogenic and Non-Pathogenic Enterococcus faecium Genomes. Journal of Genetics and Genomics, 42. https://doi.org/10.1016/j.jgg.2014.07.002
- Li, P., Gu, Q., & Zhou, Q. (2016). Complete genome sequence of Lactobacillus plantarum LZ206, a potential probiotic strain with antimicrobial activity against food-borne pathogenic microorganisms. Journal of Biotechnology, 238, 52–55. https://doi.org/10.1016/j.jbiotec.2016.09.012
- Khatri, I., Tomar, R., Ganesan, K., Prasad, G. S., & Subramanian, S. (2017). Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Scientific Reports, 7(1), 371. https://doi.org/10.1038/s41598-017-00414-2
- Morovic, W., Hibberd, A. A., Zabel, B., Barrangou, R., & Stahl, B. (2016). Genotyping by PCR and High-Throughput Sequencing of Commercial Probiotic Products Reveals Composition Biases. Frontiers in Microbiology, 7, Article 1747. https://doi.org/10.3389/fmicb.2016.01747
- Lewis, Z. T., Shani, G., Masarweh, C. F., Popovic, M., Frese, S. A., Sela, D. A., Mills, D. A. (2016). Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatric Research, 79(3), 445–452. https://doi.org/10.1038/pr.2015.244
Resources
More references can be obtained by emailing the authors or editors.








