Whole Sample Next-Generation DNA Sequencing Method: An Alternative to DNA Barcoding
This article discusses a non-targeted method for whole sample next generation DNA sequencing (NGS) that does not rely on DNA barcoding. DNA barcoding requires amplification of a specific gene region, which introduces bias. Our non-targeted method removes this bias by eliminating the amplification step. The applications of this method are broad and we have begun optimizing workflows for numerous materials, both processed and unprocessed. Some of the materials we have been able to successfully identify at the species level are fish tissue, fish meal, unrefined fish oil, unrefined plant-based oils (nuts, seeds, and fruits), specific components of cooked and processed products such as cookies and powders, and processed meats. Non-targeted NGS is also a very powerful tool to comprehensively identify constituents of microbial communities in probiotics and fermented products like kombucha. Additionally, this non-targeted technique is applicable to detection and identification of microbial contamination at various levels of manufacturing including equipment surfaces, processing water and assaying intermediate processing steps. In this communication we briefly review a current issue in the botanicals industry, discuss the methods that have been used in the past to tackle that problem, and present preliminary results from a pilot study we performed to determine the utility of non-targeted NGS in high-throughput identification of botanical raw materials.
The value of the global herbal dietary supplement (botanical) market was estimated to be greater than $90 billion in 2016, with a projected compound annual growth rate of 5-6%. Currently, regulators and manufacturers in this rapidly expanding market seek to confirm the veracity of label claims, investigate fraud, identify adulterants and ensure product quality.1 These products are often dried and ground, making visual identification difficult, time consuming and sometimes impossible.2 It is critical to this market that botanical identification be high-throughput, accurate and cost effective. Historically, various chromatography techniques have been used to meet this need, but those techniques rely on identification of molecules that can vary significantly due to storage conditions, which has led to the use of DNA barcoding as an analytical technique. However, DNA barcoding is not without significant challenges.1
For quite some time, scientists have had the ability to identify biological samples by sequencing their DNA.3 Currently DNA sequencing-based identification methods rely heavily on a technique called DNA barcoding, which functions analogously to the barcodes found on products in a grocery store. DNA barcoding amplifies a distinct small gene region that serves as a unique identifier and “scans” it by DNA sequencing.4 The advantages of this amplification are high sensitivity and simplification of data analysis. However, this amplification is not completely reliable and in practice can create biases and false positives.5 There is also the possibility that the amplification may fail, causing false negatives.6 When using DNA barcoding to identify botanical raw materials, numerous labs have observed notably high levels of apparent contamination.7 While it is certainly likely that some or even many botanical raw material samples contain contamination, it is also possible that the amplification-based method of DNA barcoding is itself contributing to the levels of contamination that are being observed.
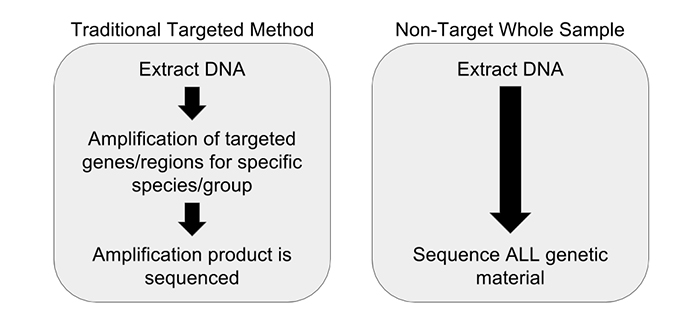
We have partnered with Practical Informatics and Pacific Northwest Genomics to develop comprehensive whole sample DNA screening methods that don’t rely on amplification. To achieve this we are utilizing a non-targeted metagenomics workflow. Non-targeted metagenomic analysis is a powerful tool for examining the entire genetic content of a sample, instead of just one particular gene region (if a gene is a word or phrase, then a genome is the entire book, and the metagenome is the library). Unlike DNA barcoding, which requires PCR amplification, non-targeted metagenomic analysis requires no prior knowledge of a sample’s source and does not introduce the biases that plague PCR initiated methods. All of the DNA extracted from a sample is analyzed without targeting any particular gene region, relying instead on complex data analysis to identify the constituents (Figure 1). This is accomplished with the use of advanced molecular biology techniques and sophisticated computational methods, combined with a highly-curated database of species-identifying DNA sequences. Our research and development team has completed several experiments demonstrating the utility of a non-targeted DNA sequencing method.
Our research endeavors to solve the issues of DNA sequence analysis that originate with the PCR step by simply eliminating amplification from our process entirely. PCR amplification as a prelude to DNA sequencing traces to traditional technologies that were lower throughput and required large amounts of material. Current generation high-throughput DNA sequencing technologies do not require large amounts of starting material, and therefore amplification can be avoided. Many DNA barcoding methods require universal primers, which, during PCR, can amplify some products but not others, leading to false negatives. A solution to that issue is to use specific primers, however this is also inherently problematic as a certain foreknowledge of the sample identity is required. What is the advantage to our non-targeted sequencing method? There is no need to direct the analysis to any particular identification before sequencing, decreasing the introduction of bias and false negatives. As an added bonus, we don’t need to know what the sample is prior to analysis—we can tell you what it is rather than you telling us.
Continue to page 2 below.
Another concern observed in PCR amplification-based barcoding is drastic over-representation of sample constituents that are actually present in quantities far below 1%, possibly even just trace amounts. This creates the perception of adulteration when there are only incidental amounts of material. This incidental DNA can originate from agricultural equipment, manufacturing surfaces or individuals handling the sample. We performed and replicated several experiments to display the advantages of a non-targeted approach over amplification-based methods. Vouchered botanical reference materials (VBRM) were used to simulate a scenario with incidental contamination, which is an area of concern.7 Chamomile (Matricaria chamomilla) and lemon balm (Melissa officinalis) were combined in a 99:1 mass ratio to show how a sample with ~1% contamination would appear to traditional DNA barcoding directly compared to our non-targeted metagenomic analysis. A ~1% threshold was used as it is less than the allowable level of background organic material (2%), according to the U.S. Pharmacopeial Convention.8 The results of these experiments were enlightening. The traditional method of DNA barcoding not only over represented the presence of the ~1% “contaminant”, but also identified numerous background incidental species (see Figure 2). In addition to the two included VRBM species, chamomile and lemon balm, eight other species were amplified by universal primers and sequenced. With the non-targeted approach, chamomile was identified with no other “contaminates” appearing in the dataset. The non-targeted method was able to determine the identity of the product without over representing incidental DNA false positives, allowing us to avoid the misinterpretation of a false positive or trace “adulterant”.
The advantage of using non-targeted whole sample sequencing is that the data are not constrained by a targeted marker. In addition to reducing bias, this opens up the possibility of analyzing mixed or composite samples as well as processed samples. Comprehensively sequencing the entirety of the DNA present in a sample also allows for the discovery of new data applications. Along with the specific goal of identifying unknown samples, this type of sequencing provides large amounts of complex data to use for further research such as biomarker prospecting. Biomarker prospecting can uncover novel genes or gene regions to use as identifiers for individuals or groups of organisms. We continue to discover innovative uses of the large datasets being generated.
References
- Mishra, P., Kumar, A., Nagireddy, A., Mani, D. N., Shukla, A. K., Tiwari, R., & Sundaresan, V. (2016). DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnology Journal, 14(1), 8–21. https://doi.org/10.1111/pbi.12419
- Baker, D. A., Stevenson, D. W., & Little, D. P. (2012). DNA barcode identification of black cohosh herbal dietary supplements. Journal of AOAC International, 95(4), 1023–1034. https://doi.org/10.5740/jaoacint.11-261
- Galimberti, A. De Mattia, F., et al. (2012). DNA barcoding as a new tool for food traceability. Food Research International 50, 55-63.
- Hebert, P. D. N., Cywinska, A., Ball, S. L., & deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings. Biological Sciences / The Royal Society, 270(1512), 313–21. https://doi.org/10.1098/rspb.2002.2218
- Taylor, H. R. & Harris, W. E. (2012). An emergent science on the brink of irrelevance: A review of the past 8 years of DNA barcoding. Molecular Ecology Resources, 12(3), 377–388. https://doi.org/10.1111/j.1755-0998.2012.03119.x
- Samarakoon, T., Wang, S. Y., & Alford, M. H. (2013). Enhancing PCR Amplification of DNA from Recalcitrant Plant Specimens Using a Trehalose-Based Additive. Applications in Plant Sciences, 1(1), 1200236. https://doi.org/10.3732/apps.1200236
- Newmaster, S., Ragupathy, S., & Hanner, Ro. (n.d.). A caution to industry and regulators – “Incidental DNA fragments” may be misinterpreted using Next Generation Sequencing (NGS).
- Sarma, N., Giancaspro, G., & Venema, J. (2016). Dietary supplements quality analysis tools from the United States Pharmacopeia. Drug Testing and Analysis, 8(3–4), 418-423. https://doi.org/10.1002/dta.1940