According to a survey by retail consulting firm Daymon Worldwide, 50% of today’s consumers are more concerned about food safety and quality than they were five years ago. Their concerns are not unfounded. Recalls are on the rise, and consumer health is put at risk by undetected cases of food adulteration and contamination.
While consumers are concerned about the quality of the food they eat, buy and sell, the brands responsible for making and selling these products also face serious consequences if their food safety programs don’t safeguard against devastating recalls.
A key cause of recalls, food fraud, or the deliberate and intentional substitution, addition, tampering or misrepresentation of food, food ingredients or food packaging, continues to be an issue for the food safety industry. According to PricewaterhouseCoopers, food fraud is estimated to be a $10–15 billion a year problem.
Some of the more notorious examples include wood shavings in Parmesan cheese, the 2013 horsemeat scandal in the United Kingdom, and Oceana’s landmark 2013 study, which revealed that a whopping 33% of seafood sold in the United States is mislabeled. While international organizations like Interpol have stepped up to tackle food fraud, which is exacerbated by the complexity of globalization, academics estimate that 4% of all food is adulterated in some way.
High-profile outbreaks due to undetected pathogens are also a serious risk for consumers and the food industry alike. The United States’ economy alone loses about $55 billion each year due to food illnesses. The World Health Organization estimates that nearly 1 in 10 people become ill every year from eating contaminated food. In 2016 alone, several high-profile outbreaks rocked the industry, harming consumers and brands alike. From the E. coli O26 outbreak at Chipotle to Salmonella in live poultry to Hepatitis A in raw scallops to the Listeria monocytogenes outbreak at Blue Bell ice cream, the food industry has dealt with many challenges on this front.
What’s Being Done?
Both food fraud and undetected contamination can cause massive, expensive and damaging recalls for brands. Each recall can cost a brand about $10 million in direct costs, and that doesn’t include the cost of brand damage and lost sales.
Frustratingly, more recalls due to food fraud and contamination are happening at a time when regulation and policy is stronger than ever. As the global food system evolves, regulatory agencies around the world are fine-tuning or overhauling their food safety systems, taking a more preventive approach.
At the core of these changes is HACCP, the long implemented and well-understood method of evaluating and controlling food safety hazards. In the United States, while HACCP is still used in some sectors, the move to FSMA is apparent in others. In many ways, 2017 is dubbed the year of FSMA compliance.
There is also the Global Food Safety Initiative (GFSI), a private industry conformance standard for certification, which was established proactively by industry to improve food safety throughout the supply chain. It is important to note that all regulatory drivers, be they public or private, work together to ensure the common goal of delivering safe food for consumers. However, more is needed to ensure that nothing slips through the food safety programs.
Now, bolstered by regulatory efforts, advancements in technology make it easier than ever to update food safety programs to better safeguard against food safety risks and recalls and to explore what’s next in food.
Powering the Food Safety Programs of Tomorrow
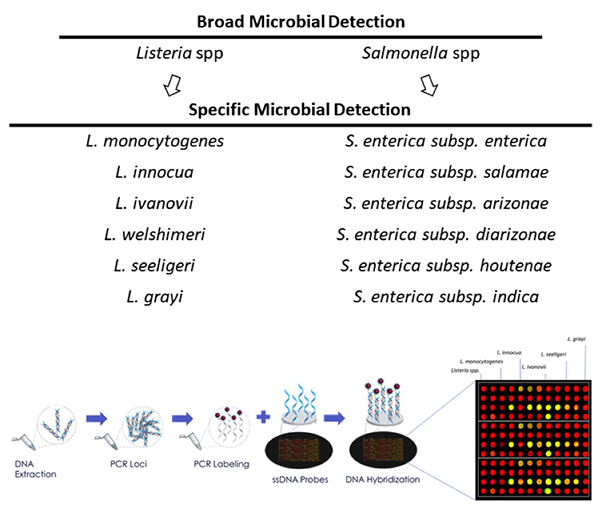
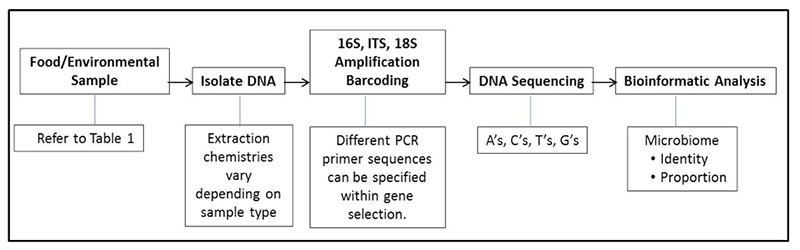
Today, food safety programs are being bolstered by new technologies as well, including genomic sequencing techniques like NGS. NGS, which stands for next-generation sequencing, is an automated DNA sequencing technology that generates and analyzes millions of sequences per run, allowing researchers to sequence, re-sequence and compare data at a rate previously not possible.
The traditional methods of polymerase chain reaction (PCR) are quickly being replaced by faster and more accurate solutions. The benefit of NGS over PCR is that PCR is targeted, meaning you have to know what you’re looking for. It is also conducted one target at a time, meaning that each target you wish to test requires a separate run. This is costly and does not scale.
Next-generation sequencing, by contrast, is universal. A single test exposes all potential threats, both expected and unexpected. From bacteria and fungi to the precise composition of ingredients in a given sample, a single NGS test guarantees that hazards cannot slip through your supply chain. In the not-too-distant future, the cost and speed of NGS will meet and then quickly surpass legacy technologies; you can expect the technology to be adopted with increasing speed the moment it becomes price-competitive with PCR.
Applications of NGS
Even today’s NGS technologies are deployment-ready for applications including food safety and supplier verification. With the bottom line protected, food brands are also able to leverage NGS to build the food chain of tomorrow, and focus funding and resources on research and development.
Safety Testing. Advances in NGS allow retailers and manufacturers to securely identify specific pathogens down to the strain level, test environmental samples, verify authenticity and ultimately reduce the risk of outbreaks or counterfeit incidents.
Compared to legacy PCR methods, brands leveraging NGS are able to test for multiple pathogens with a single test, at a lower cost and higher accuracy. This universality is key to protecting brands against all pathogens, not just the ones for which they know to look.
Supplier Verification. NGS technologies can be used to combat economically motivated food fraud and mislabeling, and verify supplier claims. Undeclared allergens are the number one reason for recalls.
As a result of FSMA, the FDA now requires food facilities to implement preventative controls to avoid food fraud, which today occurs in up to 10% of all food types. Traditional PCR-based tests cannot distinguish between closely related species and have high false-positive rates. NGS offers high-resolution, scalable testing so that you can verify suppliers and authenticate product claims, mitigating risk at every level.
R&D. NGS-based metagenomics analysis can be used in R&D and new product development to build the next-generation of health foods and nutritional products, as well as to perform competitive benchmarking and formulation consistency monitoring.
As the consumer takes more and more control over what goes into their food, brands have the opportunity to differentiate not only on transparency, but on personalization, novel approaches and better consistency.
A Brighter Future for Food Safety
With advances in genomic techniques and analysis, we are now better than ever equipped to safeguard against food safety risks, protect brands from having to issue costly recalls, and even explore the next frontier for food. As the technology gets better, faster and cheaper, we are going to experience a tectonic shift in the way we manage our food safety programs and supply chains at large.