This month we are happy to welcome our guest co-authors and interviewees Eric Brown, Ph.D. and Marc Allard, Ph.D. of CFSAN as we explore the FDA’s GenomeTrakr program in a two-part Food Genomics column. Many of our readers have heard of GenomeTrakr, but are likely to have several questions regarding its core purpose and how it will impact food producers and processors in the United States and globally. In Part I we explore some technical aspects of the topic followed by Part II dealing with practical questions.
Part I: The basics of GenomeTrakr
Greg Siragusa/Doug Marshall: Thank you Dr. Allard and Dr. Brown for joining us in our monthly series, Food Genomics, to inform our readers about GenomeTrakr. Will you begin by telling us about yourselves and your team?
Eric Brown/Marc Allard: Hello, I am Eric, the director of the Division of Microbiology at the U.S. Food and Drug Administration at the Center for Food Safety and Applied Nutrition. Our team is made up of two branches, one that specializes in developing and validating methods for getting foodborne pathogens out of many different food matrices and the other branch conducts numerous tests to subtype and characterized foodborne pathogens. The GenomeTrakr program is in the subtyping branch as Whole Genome Sequencing (WGS) is the ultimate genomic subtyping tool for characterizing a foodborne pathogen at the DNA level.
Hello, my name is Marc, I am a senior biomedical research services officer and a senior advisor in Eric’s division. We are part of the group that conceived, evaluated and deployed the GenomeTrakr database and network.
Siragusa/Marshall: Drs. Allard and Brown, imagine yourself with a group of food safety professionals ranging from vice president for food safety to director, manager and technologists. Would you please give us the ‘elevator speech’ on GenomeTrakr?
Brown/Allard: GenomeTrakr is the first of its kind distributed network for rapidly characterizing bacterial foodborne pathogens using whole genome sequences (WGS). This genomic data can help FDA with many applications, including trace-back to determine the root cause of an outbreak as well providing one work-flow for rapidly characterizing all of the pathogens for which the agency has responsibility. These same methods are also very helpful for antimicrobial resistance monitoring and characterization.
Siragusa/Marshall: From the FDA website, GenomeTrakr is described as “a distributed network of labs to utilize whole genome sequencing for pathogen identification.” We of course have very time-proven methods of microbial identification and subtyping, so why do we need GenomeTrakr for identification and subtyping of microorganisms?
Brown/Allard: If all you want to know is species identification then you are correct, there are existing methods to do this. For some applications you need full characterization through subtyping (i.e., Below the level of species to the actual strain) with WGS. WGS of pathogens provides all of the genetic information about an organism as well as any mobile elements such as phages and plasmids that may be associated with these foodborne pathogens. The GenomeTrakr network and database compiles a large amount of new genetic or DNA sequence data to more fully characterize foodborne pathogens.
GenomeTrakr and WGS are a means to track bacteria based on knowing the sequence of all DNA that comprises that specific bacterium’s genome. It can be called the “ultimate identifier” in that it will show relationships at a very deep level of accuracy.
Siragusa/Marshall: Is it an accurate statement that GenomeTrakr can be considered the new iteration of PulseNet and Pulse field gel electrophoresis (PFGE)? Will PulseNet and PFGE disappear, or will PulseNet and GenomeTrkr merge into a single entity?
Brown/Allard: PulseNet is a network of public health labs run by the CDC, with USDA and FDA as active participants. The network is alive and well and will continue subtyping pathogens for public health. The current and historical subtyping tool used by PulseNet for more than 20 years is PFGE. It is expected that CDC, USDA and FDA’s PFGE data collection will be replaced by WGS data and methods. That transformation has already begun. GenomeTrakr is a network of public health labs run by the FDA to support FDA public health and regulatory activities using WGS methods. Starting in 2012, this network is relatively new and is focused currently on using WGS for trace back to support outbreak investigations and FDA regulatory actions. CDC PulseNet has used WGS data on Listeria and collects draft genomes (i.e., unfinished versions of a final genome are used for quicker assembly) of other foodborne pathogens as well, and USDA’s FSIS has used WGS for the pathogens found on the foods that they regulate. All of the data from GenomeTrakr and Pulsenet are shared at the NCBI Pathogen Detection website (see Figure 1).
Siragusa/Marshall: Does an organism have to be classified to the species level before submitting to GenomeTrakr?
Brown/Allard: Yes, species-level identification is part of the minimal metadata (all of the descriptors related to a sample such as geographic origin, lot number, sources, ingredients etc.) required to deposit data in the GenomeTrakr database. This allows initial QA/QC metrics to determine if the new genome is labeled properly.
Siragusa/Marshall: After an isolate is identified to the species level, would you describe to the reader what the basic process is going from an isolated and speciated bacterial colony on an agar plate to a usable whole genome sequence deposited in the GenomeTrakr database?
Brown/Allard: The FDA has a branch of scientists who specialize in ways to isolate foodborne pathogens from food. The detailed methods used ultimately end up in the Bacteriological Analytical Manual (BAM) of approved and validated methods. Once a pathogen is in pure culture then DNA is extracted from the bacterial cells. The DNA is then put into a DNA sequencing library, which modifies the DNA to properly attach and run sequencing reactions depending on the specific sequencing vendor used. The sequence data is downloaded from the sequencing equipment and then uploaded to the National Center for Biotechnology Information (NCBI) Pathogen Detection website. The database is publicly open to allow anyone with foodborne pathogens to upload their data and compare their sequences to what is available in the database.
Siragusa/Marshall: Suppose a specific sequence type of a foodborne bacterial pathogen is found and identified from a processing plant but that the plant has never had a positive assay result for that pathogen in any of its history of product production and ultimate consumption. If an outbreak occurred somewhere in the world and that same specific sequence type were identified as the causative agent, would a company be in anyway liable? Could one even make an association between the two isolates with the same sequence type isolated at great distances from open another?
Brown/Allard: The genetic evidence from WGS supports the hypothesis that the two isolates shared a recent common ancestor. If, for example, the isolate from the processing plant and the outbreak sample where genetically identical across the entire genome, the prediction is that the two samples are connected in some way that is currently not understood. The genetic matches guide the FDA and help point investigations to study the possible connections. This might include additional inspection of the processing plant as well as linking this to the typical epidemiological exposure data. Sometimes due to the indirect nature of how pathogens circulate through the farm to fork continuum and the complex methods of trade, no connection is made. More commonly, these investigative leads from genetic matches help the FDA establish direct links between the two bacterial isolates through a shared ingredient, shared processing, distribution or packaging process. The genetic information and cluster helps the FDA discover new ways that the pathogens are moving from farm to fork. We are unaware of any example where identical genomes somehow independently arose and were unrelated. This is counter to molecular evolutionary theory anyway. Genetic identity equals genetic relatedness and the closer two isolates are genetically to each other, the more recent that they shared a common ancestor. With regard to liability, this is a topic beyond the scope of our group, but genomic data does not by itself prove a direct linkage and that is why additional investigations must follow any close matches.
Siragusa/Marshall: We know that SNPs (Single Nucleotide Polymorphisms or single base pair differences in the same location in a genome) are commonly used to distinguish clonality of bacteria with highly similar genomes. Are there criteria used by GenomeTrakr bioinformaticists that are set to help define what is similar, different or the same?
Brown/Allard: As the database grows with more examples of diverse serotypes or kinds of foodborne pathogens, the FDA WGS group is observing common patterns that can be used as guidance to define what is same or different. For example, closely related for Salmonella and E. coli are usually in the five or fewer SNPs, and closely related for Listeria is 20 or fewer SNPs using the current FDA validated bioinformatics pipeline. These values are not set in stone but should be considered more like guidance for what FDA and GenomeTrakr have observed already from earlier case studies that have already been collected and examined. Often, a greater number (e.g., 21-50) of SNP differences have been observed between strains isolated in some outbreaks. Any close match might support or direct an outbreak investigation if there is evidence that suggests that a particular outbreak looks most closely like an early case from a specific geographic location. WGS data helps investigators focus their efforts toward and international verses domestic exposure or possible country of origin. Even more divergent WGS linkages, when SNPs are greater than 50-100, often connect to different foods or different geographic locations that would lead investigators away from the source of an outbreak as the data provides both inclusivity as well as exclusivity.
When two strains have more than 50–100 SNPs, different food or geographic sources of those strains can be incorrectly linked resulting in investigators pursuing an incorrect source.
Siragusa/Marshall: Can SNPs be identified from different agar-plate clones of the same strain (i.e., Different colonies on the same plate)?
Brown/Allard: Since understanding the natural genetic variation present in foodborne pathogens is the basis to understanding relatedness, the FDA conducted validation experiments on growing then sequencing colonies from the same plate, colonies from frozen inocula, thawing and plating, as well as running the same DNAs on different instruments and with different sequencing technicians. The FDA’s work with Salmonella enterica Montevideo sequencing as well as ongoing proficiency testing among laboratories shows that the same isolate most often has no differences, although some samples have 1-2 SNP differences. Genetic differences observed in isolates collected by FDA inspectors all related to a common outbreak generally have more genetic differences, and this appears to be dependent on the nature of the facility and the length of time that the foodborne pathogen has been resident in the facility and the selective pressure to which the pathogen was exposed to in a range from 0–5 SNPs different.
Siragusa/Marshall: Regarding the use of WGS to track strains in a particular processing plant, is it possible that within that closed microenvironment that strains will evolve sufficiently so that it becomes unique to that source?
Brown/Allard: Yes, we have discovered multiple examples of strains that have evolved in a unique way that they appear to be specific to that source. Hospitals use the same practice to understand hospital-acquired infections and the routes of transmission within a hospitals intensive care unit or surgery. Food industry laboratories as well as FDA investigators could use WGS data in a similar way to determine the root cause of the contamination by combining WGS data with inspection and surveillance. The FDA Office of Compliance uses WGS as one piece of evidence to ask the question: Have we seen this pathogen before?
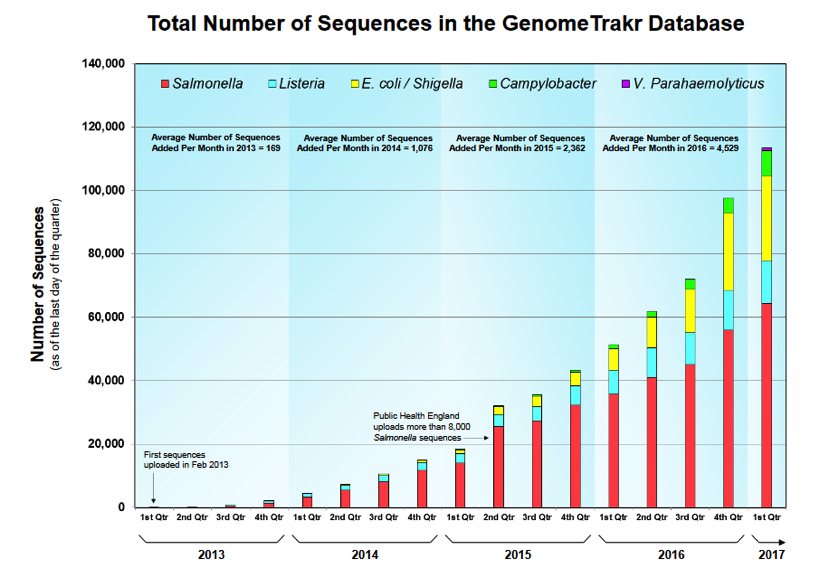
Siragusa/Marshall: The number of sequences in the GenomeTrakr database is approaching 120,000 (~4,000 per month are added). Are the sequences in the GenomeTrakr database all generated by GenomeTrakr Network labs?
Brown/Allard: The sequences labeled as GenomeTrakr isolates at the NCBI biosample and bioproject databases are the WGS efforts supported by the U.S. FDA and USDA FSIS. GenomeTrakr is a label identifying the FDA, USDA FSIS and collaborative partner’s efforts to sequence food and environmental isolates. Additional laboratories, independent and beyond formal membership in the GT network, upload WGS data to the NCBI pathogen detection website of which GenomeTrakr is one part. CDC shares WGS data on primarily clinical PulseNet isolates and USDA FSIS shares WGS foodborne pathogens for foods that they regulate. Numerous international public health laboratories also upload WGS data to NCBI. The NCBI pathogen detection website includes all publicly released WGS data for the species that they are analyzing, and this might include additional research or public health data. The point of contact for who submitted the data is listed in the biosample data sheet, an example of which can be seen here.
Siragusa/Marshall: Once sequences are deposited into the GenomeTrakr database, are they also part of GenBank?
Brown/Allard: The majority of the GenomeTrakr database is part of the NCBI SRA (sequence read archive) database, which is a less finished version of the data in GenBank. GenBank data is assembled and annotated, which takes more time and analysis to complete. Once automated software is optimized and validated, NCBI likely will place all of the GenomeTrakr data into GenBank. Currently, only the published WGS data from GenomeTrakr is available in GenBank. All of the GenomeTrakr data is available in SRA both at GenomeTrakr bioprojects and in the NCBI pathogen detection website.
Readers, look for the Part II of this column where we continue our exploration with Drs. Brown and Allard and ask some general questions about the logistics surrounding GenomeTrakr. As always, please contact either Greg Siragusa or Doug Marshall with comments, questions or ideas for future Food Genomics columns.
About the Interviewees
Marc W. Allard, Ph.D.
 Marc Allard, Ph.D. is a senior biomedical research services officer specializing in both phylogenetic analysis as well as the biochemical laboratory methods that generate the genetic information in the GenomeTrakr database, which is part of the NCBI Pathogen Detection website. Allard joined the Division of Microbiology in FDA’s Office of Regulatory Science in 2008 where he uses Whole Genome Sequencing of foodborne pathogens to identify and characterize outbreaks of bacterial strains, particularly Salmonella, E. coli, and Listeria. He obtained a B.A. from the University of Vermont, an M.S. from Texas A&M University and his Ph.D. in biology in from Harvard University. Allard was the Louis Weintraub Associate Professor of Biology at George Washington University for 14 years from 1994 to 2008. He is a Fellow of the American Academy of Microbiology.
Marc Allard, Ph.D. is a senior biomedical research services officer specializing in both phylogenetic analysis as well as the biochemical laboratory methods that generate the genetic information in the GenomeTrakr database, which is part of the NCBI Pathogen Detection website. Allard joined the Division of Microbiology in FDA’s Office of Regulatory Science in 2008 where he uses Whole Genome Sequencing of foodborne pathogens to identify and characterize outbreaks of bacterial strains, particularly Salmonella, E. coli, and Listeria. He obtained a B.A. from the University of Vermont, an M.S. from Texas A&M University and his Ph.D. in biology in from Harvard University. Allard was the Louis Weintraub Associate Professor of Biology at George Washington University for 14 years from 1994 to 2008. He is a Fellow of the American Academy of Microbiology.
Eric W. Brown, Ph.D.
 Eric W. Brown, Ph.D. currently serves as director of the Division of Microbiology in the Office of Regulatory Science. He oversees a group of 50 researchers and support scientists engaged in a multi-parameter research program to develop and apply microbiological and molecular genetic strategies for detecting, identifying, and differentiating bacterial foodborne pathogens such as Salmonella and shiga-toxin producing E. coli. Brown received his Ph.D. in microbial genetics from The Genetics Program in the Department of Biological Sciences at The George Washington University. He has conducted research in microbial evolution and microbial ecology as a research fellow in the National Cancer Institute, the U.S. Department of Agriculture, and as a tenure-track Professor of Microbiology at Loyola University of Chicago. Brown came to the Food and Drug Administration in 1999 and has since carried out numerous experiments relating to the detection, identification, and discrimination of foodborne pathogens.
Eric W. Brown, Ph.D. currently serves as director of the Division of Microbiology in the Office of Regulatory Science. He oversees a group of 50 researchers and support scientists engaged in a multi-parameter research program to develop and apply microbiological and molecular genetic strategies for detecting, identifying, and differentiating bacterial foodborne pathogens such as Salmonella and shiga-toxin producing E. coli. Brown received his Ph.D. in microbial genetics from The Genetics Program in the Department of Biological Sciences at The George Washington University. He has conducted research in microbial evolution and microbial ecology as a research fellow in the National Cancer Institute, the U.S. Department of Agriculture, and as a tenure-track Professor of Microbiology at Loyola University of Chicago. Brown came to the Food and Drug Administration in 1999 and has since carried out numerous experiments relating to the detection, identification, and discrimination of foodborne pathogens.



