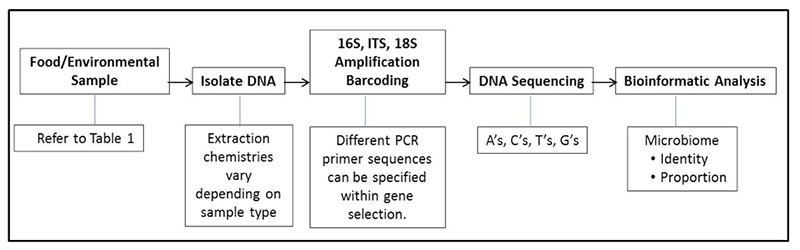
Whenever an order is placed for an aerobic plate count, lactic acid bacteria count, Enterobacteriaceae count, coliform count, fecal coliform count, Escherichia coli count, or yeast and mold count, it involves ordering an indicator test. So obviously, in food and water quality safety analyses, indicator microbiology is a highly routine and frequent activity. In fact, many business-to-business transactions are partially dictated by the outcome of indicator tests in the form of purchase specifications. Raw material producers and ingredient manufacturers are required to deliver products that meet the expectations of the buyer. Should such product exceed the predefined specifications, the expected transaction becomes nullified. Finished food product manufacturers also must meet specifications set by retailers and food service buyers. Some regulatory jurisdictions and public health agencies also are in this game, offering regulatory specifications for targeted groups of indicator microbes, such as EPA water quality specifications or FDA zero tolerance of pathogens in ready-to-eat foods. Here we argue that microbiomes can be a valuable tool to help choose and validate the best indicator(s) that may be used under a variety of circumstances.
The microbial indicator premise is that the presence or population size of a single indicator microbe or groups of microbes has some respective correlation with the presence of or population of either undesirable microbes (spoilers or pathogens) or desirable microbes (starter cultures or probiotics). On the public health side, indicator presence or populations can be used to define risk of an adverse public health outcome. Indicators also have utility in assessing process effectiveness, such as presence or populations of spore formers after a heat process. Sanitation efficacy can be judged by the amount of an appropriate indicator, such as residual ATP on a surface or presence of Listeria spp. in a floor drain. Culture houses (i.e., starter cultures, probiotics) and companies that manufacture fermented foods can do routine QC testing for the amount of metabolic byproducts (CO2 or acid development) as an indicator of microbial activity and also measure culture population levels.
Our customers frequently ask us, “Which is the best indicator for my ingredients, process, and products?” Of course they are looking for a very simple answer but the reality is, we must know many details about the ingredients, product, process, and intended use before we can offer a best guess. Clearly best guesses, even by esteemed experts, can lead to inappropriate indicator choices. At worse, standard industry practice informed by years of use may not offer appropriate scientific validation of the use of chosen indicators.
Another customer question we frequently hear: “My product is not reaching intended shelf life, but indicator counts show it should be fine. What is causing product performance failure?” In this scenario, the chosen indicator(s) may not allow for cultivation of the offending microbe, resulting in an “all clear” test result. Each indicator test (see Table I) will grow only the microbes able to multiply on the selected medium, at the selected incubation temperature, for the selected incubation time, and under the selected incubation atmosphere.1 Differences in media brand, or even slight deviations in media nutrition, media selective agents, temperature, time or atmosphere will have dramatic implications on what ultimately grows. What is telling is that there are many microbes in the sample that may not be cultivable at all, yet they may contribute to product performance failures. Wouldn’t it be nice if you could run one test and get a good snapshot of the all microbiota present in the specimen?
A further issue is that some microbes, which may be perfectly able to grow under a certain set of conditions, might be outgrown by other competitors. Therefore, they may not contribute to the countable population. If they are found as a minor population, the odds of identifying them from a plate count are remote. Such microbes may in fact contribute to product failure and yet never be detected by an indicator assay.
An example of well-publicized historical misuse of indicators is the application of fecal coliform counts as indicators of fecal contamination of some dry leafy food products, such as tea leaves. For decades, periodic popular press exposé articles about food service iced tea with high fecal coliform counts have appeared in the news. The respective author’s dramatic conclusion is that such teas are contaminated with feces and a threat to public health. However, in reality, when the bacteria associated with these high counts were actually identified, they were determined to be natural constituents of dry tea leaves and had no association with animal feces.2
An unconventional hypothetical indicator example seems worthy here. If you are a manufacturer of a dried ready-to-eat product or ingredient and your hazard analysis has identified Salmonella as a reasonably foreseeable environmental hazard, most will choose coliform, fecal coliform, E. coli, and/or Enterobacteriaceae counts as potential indicators. We’re sure this sounds familiar so you’re feeling pretty good about now—well, read on, please. What may be less obvious is the potential usefulness of a yeast and mold count for this purpose, because low-level moisture intrusion may lead to growth of these fungal groups and also may lead to enhanced survival/growth of Salmonella. Therefore, one may find the best indicator by looking for an indicator of moisture control problems rather than an indicator of potential fecal contamination.
Finally, verification screening of all raw materials, ingredients, processes, environmental locations, and products using traditional microbiology tests can quickly become expensive if you are looking at all the potential indicators shown in the table. By first running a single microbiome on a specimen, the predominant microbes and their relative proportional populations will be determined. This knowledge can be used to develop appropriate targeted verification screening for indicators that you now know are relevant to the specimen. Furthermore, the impact of changes in suppliers, processes, or product formulation can be measured using microbiomes to again gain confidence that appropriate indicators are still being used.
We hope this installment of Food Genomics triggers the reader to rethink the indicators they are using and ask the following questions:
- Why are we using our chosen indicators?
- Are our indicators telling us what we really need to know?
- Are there better indicators for my supplier verification program?
- Are there better indicators for my process verification program?
- Are there better indicators for my environmental monitoring program?
- Are there better indicators that more accurately predict product shelf life?
| Indicator Test |
Uses |
Microbiome |
| Aerobic Mesophilic Plate Count |
Estimate population of microbes able to grow at 35°C with air. Overall food quality indicator, shelf life/spoilage predictor |
Names of predominant microbes and relative proportions of each constituting the aerobic mesophilic population |
| Anaerobic Mesophilic Plate Count |
Estimate populations of microbes able to grow at 35°C without oxygen. Shelf life/spoilage predictor of vacuum packaged or modified atmosphere packaged foods. |
Names of predominant microbes and relative populations of each constituting the anaerobic mesophilic population |
| Standard Plate Count |
Similar to APC but used for dairy products. Estimate population of microbes able to grow at 30°C with air. Overall milk quality indicator, shelf life/spoilage predictor |
Names of predominant microbes and relative populations of each constituting the aerobic mesophilic population |
| Psychrotrophic Plate Count |
Estimate population of microbes able to grow at refrigerated temperatures (incubation temperature can vary from 5° to 15°C) with air. Shelf life/spoilage predictor |
Names of predominant microbes and relative populations of each constituting the aerobic psychrotrophic population |
| Anaerobic Psychrotrophic Plate Count |
Estimate population of microbes able to grow at refrigerated temperatures without oxygen. Refrigerated shelf life/spoilage predictor of vacuum or modified atmosphere packaged foods |
Names of predominant microbes and relative proportions of each constituting the anaerobic psychrotrophic population |
| Aerobic Thermophilic Plate Count |
Estimate population of bacterial spores able to grow at high storage temperatures (incubation temperature can vary but usually >45°C) in air or survive a thermal process. Indicator of process failure. Spoilage indicator of improperly hot held foods |
Names of predominant spores and relative proportions of each constituting the aerobic thermophilic population |
| Aerobic Mesophilic Spore |
Estimate population of bacterial spores able to grow at 35°C with air. May indicate possible Bacillus cereus risk. |
Names of predominant spores and relative proportions of each constituting the aerobic mesophilic spore population |
| Anaerobic Mesophilic Spore Count |
Estimate population of bacterial spores able to grow at 35°C without oxygen. Potential shelf life/spoilage indicator of vacuum or modified atmosphere packaged foods. May indicate possible Clostridium botulinum risk. |
Names of predominant spores and relative proportions of each constituting the anaerobic spore population |
| Aerobic Psychrophilic Spore Count |
Estimate population of bacterial spores able to grow at refrigeration temperature with air. Spoilage indicator of refrigerated foods |
Names of predominant spores and relative proportions of each constituting the aerobic psychrotrophic spore population |
| Anaerobic Psychrophilic Spore Count |
Estimate population of bacterial spores able to grow at refrigeration temperature without oxygen. Potential shelf life/spoilage indicator of refrigerated vacuum or modified atmosphere packaged foods. May indicate possible nonproteolytic Clostridium botulinum risk |
Names of predominant spores and relative proportions of each constituting the anaerobic psychrotrophic spore population |
| Aerobic Thermophilic Spore Count |
Estimate population of bacterial spores able to grow at high temperature in air. Spoilage indicator of heat processed foods. |
Names of predominant spores and relative proportions of each constituting the aerobic thermophilic spore population |
| Anaerobic Thermophilic Spore Count |
Estimate population of bacterial spores able to grow at high temperature without oxygen. Spoilage indicator of heat processed, vacuum or modified atmosphere packaged foods |
Names of predominant spores and relative proportions of each constituting the anaerobic thermophilic spore population |
| Thermoduric Plate Count |
Estimate population of microbes able to survive a pasteurization process. Used as a shelf life/spoilage predictor |
Names of predominant microbes and relative proportions surviving a thermal process |
| Lactic Acid Bacteria Count |
Estimate population of bacteria able to produce lactic acid during growth. Indicator of fermentation success or spoilage failure |
Names of predominant microbes and relative proportions that produce lactic acid |
| Proteolytic Plate Count |
Estimate population of microorganisms that produce protease enzymes. Indicator of putrefactive spoilage potential |
Names of predominant microbes and relative proportions that produce proteases |
| Lipolytic Plate Count |
Estimate population of microorganisms that produce lipase enzymes. Indicator of lipid hydrolytic rancidity spoilage potential |
Names of predominant microbes and relative proportions that produce lipases |
| Saccharolytic Plate Count |
Estimate population of microorganisms that produce amylase enzymes. Indicator of starch hydrolysis spoilage potential |
Names of predominant microbes and relative proportions that produce amylases |
| Pectinolytic Plate Count |
Estimate population of microorganisms that produce pectinase enzymes. Indicator of pectin hydrolysis spoilage potential |
Names of predominant microbes and relative proportions that produce pectinases |
| Aciduric Plate Count |
Estimate population of microorganisms able to grow in a high acid/low pH food. Indicator of spoilage potential |
Names of predominant microbes and relative proportions surviving an a high acid product |
| Aciduric Flat Sour Sporeformer Count |
Estimate population of bacterial spores able to tolerate high acid foods and produce acid without gas production. Indicator of high-acid canned food spoilage potential |
Names of predominant bacterial spores and relative proportions that grow in a high-acid canned food |
| Thermophilic Flat Sour Spore Former Count |
Estimate population of bacterial spores able to grow at high temperature and produce acid. Indicator of canned food spoilage potential |
Names of predominant bacterial spores and relative proportions that grow and produce acid in a canned food |
| Sulfide Sporeformer Count |
Estimate populations of bacterial spores that produce sulfur aroma compounds. Indicator of canned food spoilage potential |
Names of predominant bacterial spores that produce sulfur compounds |
| Halophilic Plate Count |
Estimate population of microorganisms able to grow at high salt concentrations. Indicator of microbes that can spoil low water activity foods |
Names of predominant microbes and relative proportions that grow in a high-salt food |
| Osmophilic Plate Count |
Estimate population of microorganisms able to grow at high sugar concentrations. Indicator of microbes that can spoil low water activity foods |
Names of predominant microbes and relative proportions that grow in a high-sugar food |
| Yeast & Mold Count |
Estimate population of fungal microbes. Indicator of fermentation success (mold-ripened cheeses) or spoilage potential |
Names of predominant fungi and relative proportions in a specimen |
| Preservative Resistant Yeast & Mold Count |
Estimate population of fungi able to grow or survive in the presence of a food preservative. Indicator of spoilage potential |
Names of predominant fungi and relative proportions that grow in the presence of a food preservative |
| Coliform Count |
Estimate population of bacteria able to ferment lactose at 35°C within 48 hours with gas production. Indictor of sanitation failure or possible presence of fecal pathogens |
Names of predominant bacteria and relative proportions constituting the coliform population of a specimen |
| Fecal Coliform Count |
Estimate population of bacteria able to ferment lactose at 44°C within 48 hours with gas production. Indictor of possible presence of fecal pathogens |
Names of predominant bacteria and relative proportions constituting the fecal coliform population of a specimen |
| E. coli count |
Estimate population of Escherichia coli. Indicator of fecal contamination and possible presence of enteric pathogens |
A microbiome is not a useful addition for this species-specific test |
| Total Enterobacteriaceae Count |
Estimate population of bacteria able to ferment glucose at 35°C within 24 hours. Indictor of sanitation failure or possible presence of fecal pathogens |
Names of predominant bacteria and relative proportions constituting the Enterobacteriaceae population of a specimen |
| Enterococcus Plate Count |
Estimate population of enterococci. Indicator of possible fecal contamination |
Names of predominant bacteria and relative proportions constituting the enterococci population of a specimen |
| Listeria spp. |
Estimate of the presence/absence of Listeria species in a food or environmental sample. Indicator of the possible presence of the pathogen Listeria monocytogenes |
Names of predominant Listeria species and relative proportions constituting the Listeria population of a specimen |
| Relative ATP Concentration |
Estimate the amount of adenosine triphosphate in a food or environmental sample. Indicator for the presence of living cells (food or microbial). Used as an indicator of proper sanitation |
Names of predominant microorganisms and relative proportions constituting the microbial population of a specimen taken at the same site swabbed for ATP |
| Table I. Common microbial indicator tests and the use of microbiomes for validation of effectiveness.1 |
References
- Salfinger, Y, and M.L. Tortorello. (2015). Compendium of Methods for the Microbiological Examination of Foods, 5th Ed. American Public Health Association, Washington, D.C.
- Zhao, T., M.R.S. Clavero, M. Doyle, and L.R. Beuchat. (1997). Health relevance of the presence of fecal coliforms in iced tea and leaf tea. J. Food Prot. 60(3):215-218.