To cap off a tumultuous year for foodborne illnesses, the end of 2018 saw a rather large E. coli outbreak that affected several different types of lettuce. In all, about 62 people got sick in the United States, with another 29 affected in Canada. The outbreak was traced back to a farm in California thanks to a specific DNA fingerprint in the E. coli. It started in a water reservoir and spread to the nearby crops.
Unfortunately, the event was only one of two separate incidents involving romaine lettuce last year. Another E.coli outbreak was traced back to a source in Arizona. Are these outbreaks more common than we realize? The CDC estimates that 48 million Americans fall ill each year from foodborne pathogens. Of those who get sick, 128,000 have to be hospitalized, and about 3,000 perish.
It’s clear that the industry as a whole needs to buckle down and find more effective solutions, not just for preventing outbreaks but also for mitigating damage when they happen. A new level of safety and management can be achieved with the help of many new, innovative technologies.
The following are some of the technology tools shaping the future of food safety and quality management fields.
Blockchain
As a result of the E. coli outbreak, Walmart implemented blockchain technology to track leafy greens and boost supply chain transparency. The systems and infrastructure is anticipated to be in place by the end of 2019.
Blockchain is a secure, digital ledger. It holds information about various transactions and data, all of which are carried out on the network. It’s called a blockchain because each data set within the network is a chunk or “block,” and they’re all linked to one another—hence the chain portion of the name. What this allows for is complete transparency throughout the supply chain, because you can track goods from their origin all the way to distribution and sale.
Each block is essentially a chunk of information, and when it’s entered into the chain, it cannot be altered, modified or manipulated. It’s simply there for viewing publicly. You cannot alter information contained within a single block without modifying the entire chain—which operates much like a peer-to-peer network and is split across many devices and servers.
This unique form of security establishes trust, accuracy and a clear representation of what’s happening. It allows a company to track contaminated foods along their journey, stopping them before they contaminate other goods or reach customers.
Infrared Heating
Thanks to the rising popularity of ready-to-eat meals, the industry is under pressure to adopt preservation and pasteurization methods. Particularly, they must be able to sanitize foods and package them with minimal exposure and bacteria levels. This practice allows them to stay fresh for longer and protects customers from potential foodborne illness.
Infrared heating is a method of surface pasteurization, and has been used for meats such as ham. Infrared lamps radiate heat at low temperatures, effectively killing surface bacteria and contaminants. The idea is to decontaminate or sanitize the surface of foods before final packaging occurs.
Industrial IoT and Smart Sensors
The food and beverage industry has a rather unique challenge with regard to supply chain operations. Food may be clean and correctly handled at the source with no traces of contamination, but it’s then passed on to a third party, which changes the game. Maybe a refrigerated transport breaks down, and the food within is thawed out. Perhaps a distributor doesn’t appropriately store perishable goods, resulting in serious contamination.
This transportation stage can be more effectively tracked and optimized with the help of modern IoT and smart, connected sensors. RFID tags, for instance, can be embedded in the packaging of foods to track their movements and various stats. Additional sensors can monitor storage temps, travel times, unexpected exposure, package tears and more.
More importantly, they’re often connected to a central data processing system where AI and machine learning platforms or human laborers can identify problematic changes. This setup allows supply chain participants to take action sooner in order to remedy potential problems or even pull contaminated goods out of the supply.
They can also help cut down on fraud or falsified records, which is a growing problem in the industry. Imagine an event where an employee says that a package was handled properly via forms or reporting tools, yet it was exposed to damaging elements. The implications of even simple fraud can be significant. Technology that automatically and consistently reports information—over manual entry—can help eliminate this possibility altogether.
Next-Generation Sequencing
NGS refers to a high-throughput DNA sequencing process that is now available to the food industry as a whole. It’s cheaper, more effective and takes a lot less time to complete, which means DNA and RNA sequencing is more accessible to food companies and suppliers now than it ever has been.
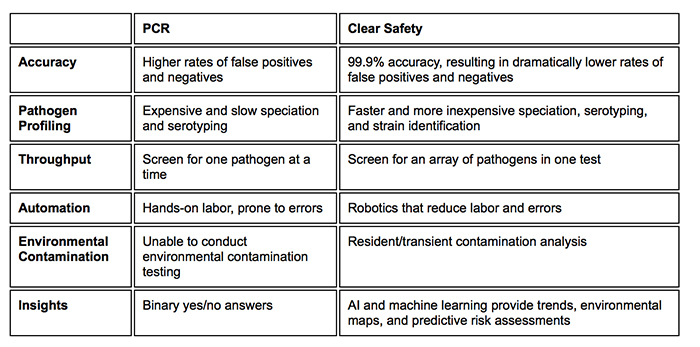
NGS can be used to assess and sequence hundreds of different samples at a time at rates of up to 25 million reads per experiment. What that means is that monitoring teams can accurately identify foodborne pathogens and contamination at the speed of the modern market. It is also a highly capable form of food safety measurement and is quickly replacing older, molecular-based methods like PCR.
Ultimately, NGS will lead to vastly improved testing and measurement processes, which can identify potential issues faster and in higher quantities than traditional methods. The food industry will be all the better and safer for it.
The Market Is Ever Evolving
While these technologies are certainly making a splash—and will shape the future of the food safety industry—they do not exist in a vacuum. There are dozens of other technologies and solutions being explored. It is important to understand that many new technologies could rise to the surface even within the next year.
The good news is that it’s all meant to improve the industry, particularly when it comes to the freshness, quality and health of the goods that consumers eat.