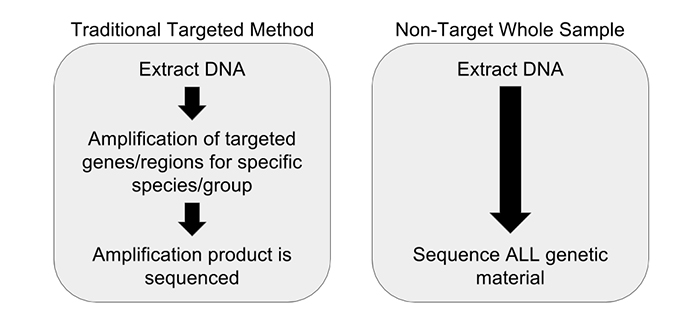
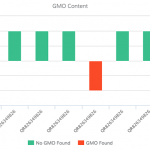
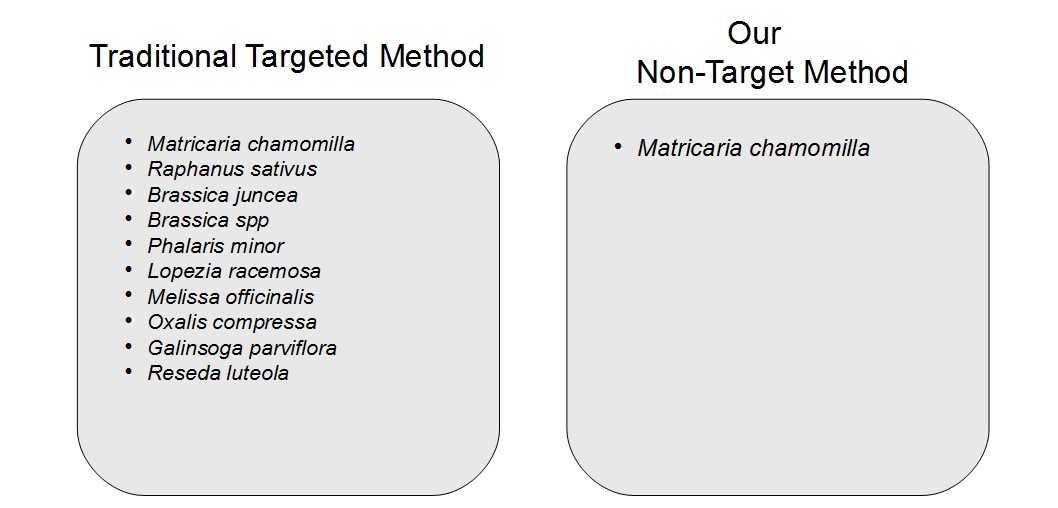
Another concern observed in PCR amplification-based barcoding is drastic over-representation of sample constituents that are actually present in quantities far below 1%, possibly even just trace amounts. This creates the perception of adulteration when there are only incidental amounts of material. This incidental DNA can originate from agricultural equipment, manufacturing surfaces or individuals handling the sample. We performed and replicated several experiments to display the advantages of a non-targeted approach over amplification-based methods. Vouchered botanical reference materials (VBRM) were used to simulate a scenario with incidental contamination, which is an area of concern.7 Chamomile (Matricaria chamomilla) and lemon balm (Melissa officinalis) were combined in a 99:1 mass ratio to show how a sample with ~1% contamination would appear to traditional DNA barcoding directly compared to our non-targeted metagenomic analysis. A ~1% threshold was used as it is less than the allowable level of background organic material (2%), according to the U.S. Pharmacopeial Convention.8 The results of these experiments were enlightening. The traditional method of DNA barcoding not only over represented the presence of the ~1% “contaminant”, but also identified numerous background incidental species (see Figure 2). In addition to the two included VRBM species, chamomile and lemon balm, eight other species were amplified by universal primers and sequenced. With the non-targeted approach, chamomile was identified with no other “contaminates” appearing in the dataset. The non-targeted method was able to determine the identity of the product without over representing incidental DNA false positives, allowing us to avoid the misinterpretation of a false positive or trace “adulterant”.
The advantage of using non-targeted whole sample sequencing is that the data are not constrained by a targeted marker. In addition to reducing bias, this opens up the possibility of analyzing mixed or composite samples as well as processed samples. Comprehensively sequencing the entirety of the DNA present in a sample also allows for the discovery of new data applications. Along with the specific goal of identifying unknown samples, this type of sequencing provides large amounts of complex data to use for further research such as biomarker prospecting. Biomarker prospecting can uncover novel genes or gene regions to use as identifiers for individuals or groups of organisms. We continue to discover innovative uses of the large datasets being generated.
References
- Mishra, P., Kumar, A., Nagireddy, A., Mani, D. N., Shukla, A. K., Tiwari, R., & Sundaresan, V. (2016). DNA barcoding: An efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnology Journal, 14(1), 8–21. https://doi.org/10.1111/pbi.12419
- Baker, D. A., Stevenson, D. W., & Little, D. P. (2012). DNA barcode identification of black cohosh herbal dietary supplements. Journal of AOAC International, 95(4), 1023–1034. https://doi.org/10.5740/jaoacint.11-261
- Galimberti, A. De Mattia, F., et al. (2012). DNA barcoding as a new tool for food traceability. Food Research International 50, 55-63.
- Hebert, P. D. N., Cywinska, A., Ball, S. L., & deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proceedings. Biological Sciences / The Royal Society, 270(1512), 313–21. https://doi.org/10.1098/rspb.2002.2218
- Taylor, H. R. & Harris, W. E. (2012). An emergent science on the brink of irrelevance: A review of the past 8 years of DNA barcoding. Molecular Ecology Resources, 12(3), 377–388. https://doi.org/10.1111/j.1755-0998.2012.03119.x
- Samarakoon, T., Wang, S. Y., & Alford, M. H. (2013). Enhancing PCR Amplification of DNA from Recalcitrant Plant Specimens Using a Trehalose-Based Additive. Applications in Plant Sciences, 1(1), 1200236. https://doi.org/10.3732/apps.1200236
- Newmaster, S., Ragupathy, S., & Hanner, Ro. (n.d.). A caution to industry and regulators – “Incidental DNA fragments” may be misinterpreted using Next Generation Sequencing (NGS).
- Sarma, N., Giancaspro, G., & Venema, J. (2016). Dietary supplements quality analysis tools from the United States Pharmacopeia. Drug Testing and Analysis, 8(3–4), 418-423. https://doi.org/10.1002/dta.1940