In a novel large-scale collaboration by IBM and Mars, Inc., researchers are harvesting and sequencing the DNA and RNA of simple food samples to determine where anomaly and mutations occur when paired with common organisms or genes, toxins, and heavy metals.
Resulting in a “microbial baseline,” or a benchmark representing normal microbe communities, the index produced from this study will be a gold standard for food and health officials globally to understand what triggers contamination and the spread of disease.
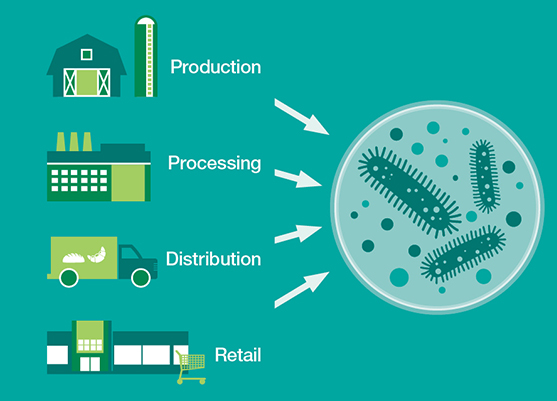
This Consortium for sequencing the food supply chain will study the microbial ecology of foods and their processing environments, and hopes to have a deeper understanding of the populations in these ecologies — bacteria, fungi, viruses, and other microorganisms — and how they interact with each other can then be used to develop new methods for keeping food safe.
“Investigating the genetic fingerprints of food ingredients and their environments will help us unearth genomic keys to healthy food and people,” describes Jeff Wesler, Vice President and Lab Director for IBM Research.
 One of the goals of the consortium is to see if there are any actions that food producers can take in respect to microbiomes that can reduce risk and make production safer. And Mars, with more than 130 factories worldwide, can help map the flow of microorganisms into and through the supply chain on a global level. An informatics infrastructure developed in the IBM Accelerated Discovery Lab, a data and analytics hub for IBM researchers and their clients and partners, will help the team parse and aggregate terabytes of genomic data from Mars and apply decades of refined analytics to uncover new insights. Adding relevant weather, transport and other contextual data could help define a targeted breakout, marking on the index a warning for food producers and distributors at the outset of a processing cycle.
One of the goals of the consortium is to see if there are any actions that food producers can take in respect to microbiomes that can reduce risk and make production safer. And Mars, with more than 130 factories worldwide, can help map the flow of microorganisms into and through the supply chain on a global level. An informatics infrastructure developed in the IBM Accelerated Discovery Lab, a data and analytics hub for IBM researchers and their clients and partners, will help the team parse and aggregate terabytes of genomic data from Mars and apply decades of refined analytics to uncover new insights. Adding relevant weather, transport and other contextual data could help define a targeted breakout, marking on the index a warning for food producers and distributors at the outset of a processing cycle.
Mars and IBM are looking to partner with industry, academics, regulators and some NGOs on the project. For more information, click here.