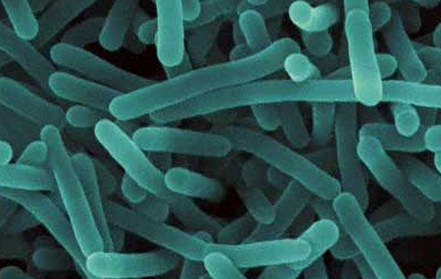
USDA announced that the agency has awarded bioMérieux’s Gene-Up® Campylobacter as the method of
choice for Campylobacter Detection in USDA FSIS Labs. GENE-UP® CAMPYLOBACTER is a real-time PCR-based solution that delivers results in under an hour and is AOAC validated for a variety of enrichment medias.
“We are thrilled to be awarded this contract by the USDA-FSIS. With the inclusion of GENE-UP® CAMPYLOBACTER, bioMérieux now has the most FSIS methods of choice for microbiology than any other diagnostics provider,” says Miguel Villa, Sr. Vice President, Americas, Industrial Applications, at bioMérieux. “This is a testament to our continued dedication to providing innovation within the animal protein sector and the food industry at large, reinforcing GENE-UP® as the full solution for all molecular testing needs in one place.”
The GENE-UP® CAMPYLOBACTER assay is the latest of bioMérieux’s molecular diagnostic solutions to be recognized by food safety regulatory bodies in the United States. Earlier this year, bioMérieux announced a research collaboration to improve microbial detection of food-borne pathogens with the Food and Drug Administration, and in 2022,
the USDA named bioMérieux’s GENE-UP® QUANT Salmonella quantification method of choice in all FSIS laboratories, with both GENE-UP® QUANT Salmonella and TEMPO® solutions currently included in the USDA Microbiology Laboratory Guidebook (MLG).
“Our Augmented Diagnostics approach helps food industry leaders go beyond the test results and harness data, genomics, and diagnostic insights to solve the toughest problems,” says John Shultz, Sr. Director, Sales and Marketing, Industrial Applications, at bioMérieux. “The recognition of GENE-UP® CAMPYLOBACTER as the method of choice of the USDA is yet another way that bioMérieux helps facilitate and advance the science of food safety and protect public health.”